Antoine Simon, now a Ph.D. student at the University of Liège, Belgium, published his assembly and study of the structure of the mitochondrial genome in a lichen forming fungus: Simon, A., Y. Liu, E. Sérusiaux & B. Goffinet. 2017. Complete mitogenome sequence of Ricasolia amplissima (Lobariaceae) reveals extensive mitochondrial DNA rearrangement within the Peltigerales (lichenized ascomycetes). The Bryologist 120(3): 335–339. pdf
Antoine initiated this study while visiting UConn in 2015, and completed it during his stay last spring. The voucher and other collections made by Antoine are deposited in the CONN herbarium.
Abstract reads: The structure of mitochondrial genomes varies among non-lichenized fungi in terms of their genic and intronic content and genic order. Whether lichenized fungal mitogenomes are equally labile is unknown due to the paucity of available mitogenomes. We assembled the mitogenome of Ricasolia amplissima (Peltigerales, Lobariaceae), using massive parallel sequencing, and compared its structure to that of two species of Peltigera (Peltigeraceae). The mitochondrial genome of R. amplissima comprised 82,333 bp, with a 29.8% G+C content, and holds 15 unique protein-coding genes, 29 tRNA genes, two rRNA genes, and one non-coding RNA gene. Although the protein-coding gene content in the mitogenome of Peltigera and Ricasolia was identical, the relative gene order differed substantially, revealing that significant gene rearrangements also characterize the evolution of mitogenomes of lichenized ascomycetes at a relatively shallow phylogenetic depth, such as within the order Peltigerales.

 In a recent article entitled “
In a recent article entitled “

 Drs. Janine Caira and Kirsten Jensen edited a special volume of the University of Kansas, Museum of Natural History publication entitled “
Drs. Janine Caira and Kirsten Jensen edited a special volume of the University of Kansas, Museum of Natural History publication entitled “
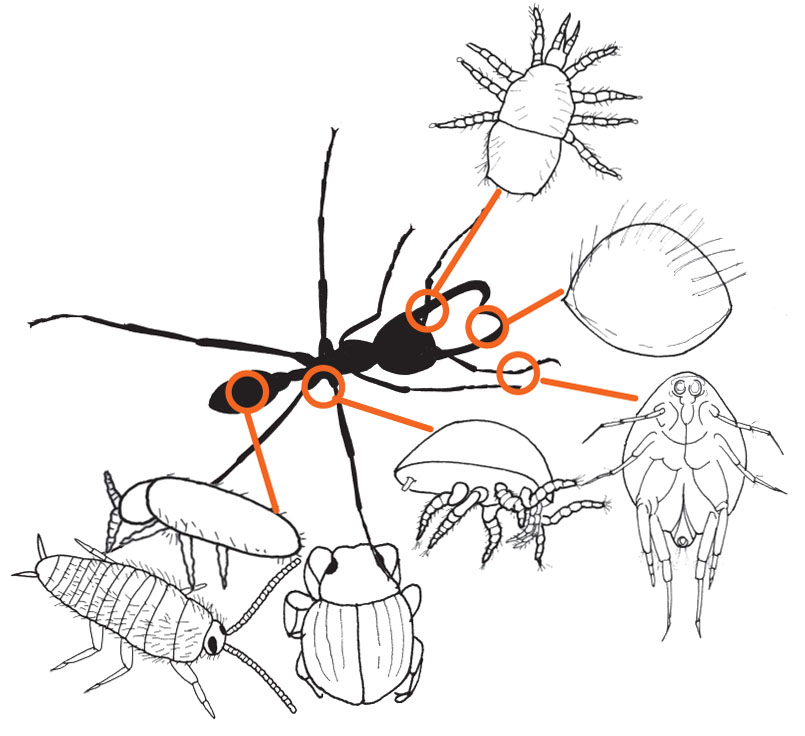 The exhibit on the life of army ants and their guests has opened in the Biology/Physics building (see
The exhibit on the life of army ants and their guests has opened in the Biology/Physics building (see