Vuruputoor V.S., A. Starovoitov, Y. Cai, Y. Liu, N. Rahmatpour, T. Hedderson, N. Wilding, J.L. Wegrzyn & B. Goffinet. 2024. Crossroads of assembling a moss genome: navigating contaminants and horizontal gene transfer in the moss Physcomitrellopsis africana. G3: Genes, Genomes, Genetics 14(7): jkae104. pdf
Abstract reads: The first chromosome-scale reference genome of the rare narrow-endemic African moss Physcomitrellopsis africana (P. africana) is presented here. Assembled from 73 × Oxford Nanopore Technologies (ONT) long reads and 163 × Beijing Genomics Institute (BGI)-seq short reads, the 414 Mb reference comprises 26 chromosomes and 22,925 protein-coding genes [Benchmarking Universal Single-Copy Ortholog (BUSCO) scores: C:94.8% (D:13.9%)]. This genome holds 2 genes that withstood rigorous filtration of microbial contaminants, have no homolog in other land plants, and are thus interpreted as resulting from 2 unique horizontal gene transfers (HGTs) from microbes. Further, P. africana shares 176 of the 273 published HGT candidates identified in Physcomitrium patens (P. patens), but lacks 98 of these, highlighting that perhaps as many as 91 genes were acquired in P. patens in the last 40 million years following its divergence from its common ancestor with P. africana. These observations suggest rather continuous gene gains via HGT followed by potential losses during the diversification of the Funariaceae. Our findings showcase both dynamic flux in plant HGTs over evolutionarily “short” timescales, alongside enduring impacts of successful integrations, like those still functionally maintained in extant P. africana. Furthermore, this study describes the informatic processes employed to distinguish contaminants from candidate HGT events.
 Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023 Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227.
Zhang L., Q. Zuo, W.Z. Ma, J. Shevock, N. Patel, M. Johnson, R. Medina, N. Wilding & B. Goffinet. 2023 Phylogenomics resolves the rediscovered Himalayan endemic Brachymeniopsis gymnostoma (Bryophyta, Funariaceae), as a species of Entosthodon, prompting also the transfer of Clavitheca poeltii. Taxon 72: 1216–1227. 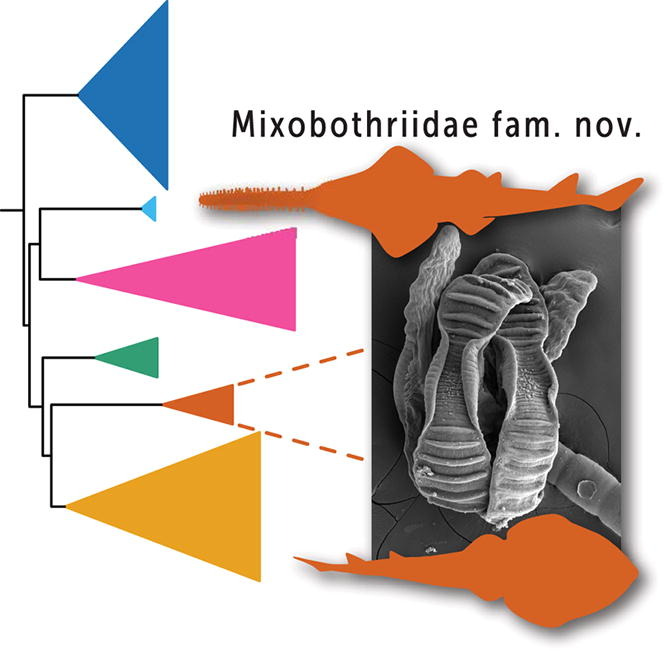 Herzog, K.S., J.N. Caira, P.Kumar Kar & K. Jensen. 2023. Novelty and phylogenetic affinities of a new family of tapeworms (Cestoda: Rhinebothriidea) from endangered sawfish and guitarfish. International Journal for Parasitology 53: 347–362.
Herzog, K.S., J.N. Caira, P.Kumar Kar & K. Jensen. 2023. Novelty and phylogenetic affinities of a new family of tapeworms (Cestoda: Rhinebothriidea) from endangered sawfish and guitarfish. International Journal for Parasitology 53: 347–362. 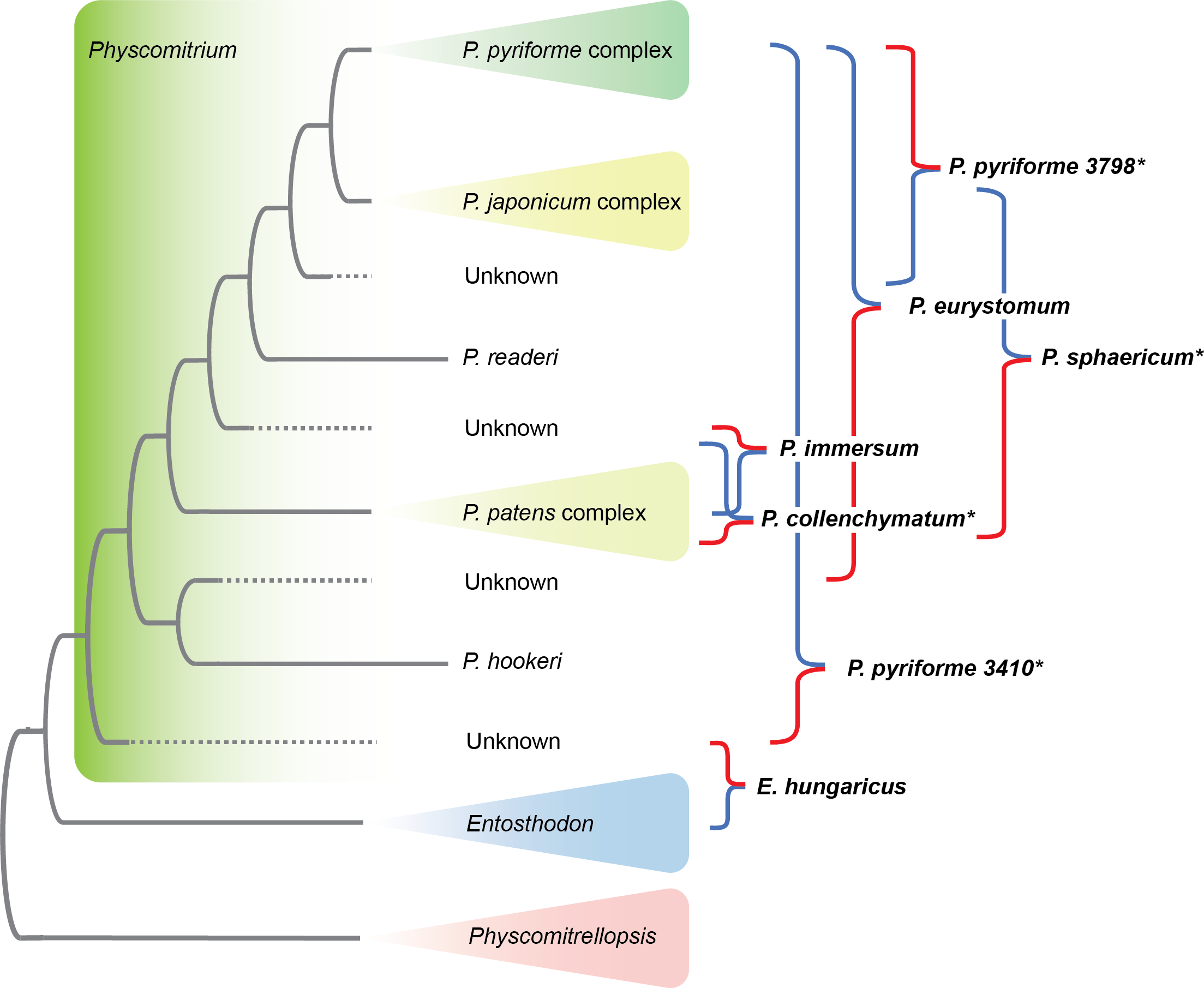 Patel N., R. Medina, L.D. Williams, O. Lemieux, B. Goffinet & M.G. Johnson. Frequent allopolyploidy with distant progenitors in the moss genera Physcomitrium and Entosthodon (Funariaceae) identified via subgenome phasing of targeted nuclear genes.
Patel N., R. Medina, L.D. Williams, O. Lemieux, B. Goffinet & M.G. Johnson. Frequent allopolyploidy with distant progenitors in the moss genera Physcomitrium and Entosthodon (Funariaceae) identified via subgenome phasing of targeted nuclear genes. 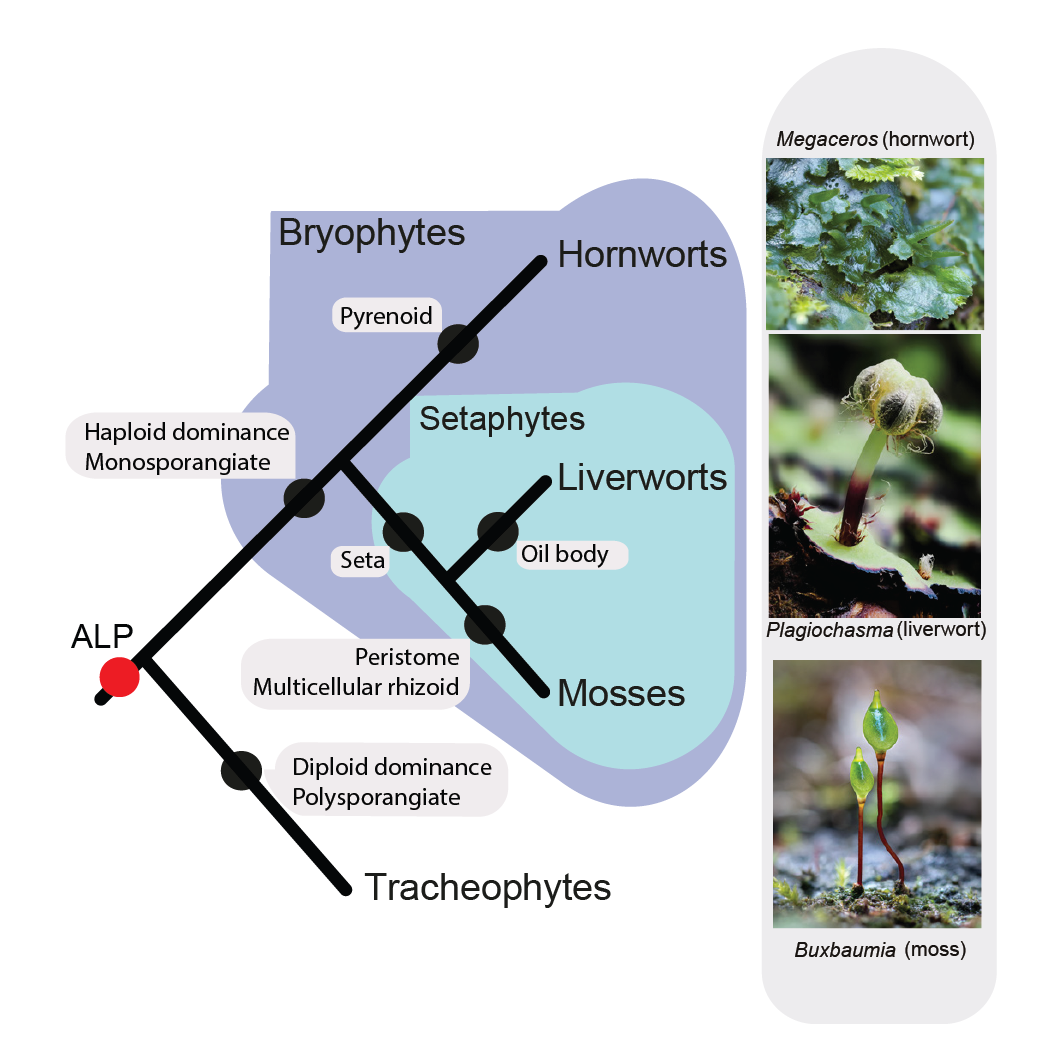 Bechteler J., G. Peñaloza-Bojacá, D. Bell, G. Burleigh, S. F. McDaniel, E. C. Davis, E. B. Sessa, A. Bippus, D. Christine Cargill, S. Chantanoarrapint, I. Draper, L. Endara, L. L. Forrest, R. Garilleti, S. W. Graham, S. Huttunen, J. Jauregui Lazo, F. Lara, J. Larraín, L. R. Lewis, D. G. Long, D. Quandt, K. Renzaglia, A. Schäfer-Verwimp, G. Ee Lee, A. S. Pinilla, M. von Konrat, C. E. Zartman, M. Regina Pereira, B. Goffinet & J. C. Villarreal. 2023. Comprehensive phylogenomic time tree of bryophytes reveals deep relationships and uncovers gene incongruences in the last 500 million years of diversification. American Journal of Botany 110: e16249.
Bechteler J., G. Peñaloza-Bojacá, D. Bell, G. Burleigh, S. F. McDaniel, E. C. Davis, E. B. Sessa, A. Bippus, D. Christine Cargill, S. Chantanoarrapint, I. Draper, L. Endara, L. L. Forrest, R. Garilleti, S. W. Graham, S. Huttunen, J. Jauregui Lazo, F. Lara, J. Larraín, L. R. Lewis, D. G. Long, D. Quandt, K. Renzaglia, A. Schäfer-Verwimp, G. Ee Lee, A. S. Pinilla, M. von Konrat, C. E. Zartman, M. Regina Pereira, B. Goffinet & J. C. Villarreal. 2023. Comprehensive phylogenomic time tree of bryophytes reveals deep relationships and uncovers gene incongruences in the last 500 million years of diversification. American Journal of Botany 110: e16249.